Poster Presentation 21st International Conference on Biological Inorganic Chemistry 2025
Emergence of catalysis from a designed generalist binding protein (#552)
The evolution of proteins that bind to small molecules and catalyze chemical transformations played a central role in the emergence of life. While natural proteins have finely tuned affinity for their primary ligands, they also have weak affinities for other molecules, which serve as starting points for the evolution of entirely new specificities. Inspired by this concept, we determined the evolvability for new functionalities of a previously designed de novo protein ABLE, which specifically binds to the drug apixaban. We showed that ABLE binds with low affinity to a subset of a diverse array of small molecules (< 300 Da) by crystallographic fragment screening. This information was further utilized to design a highly efficient, specialist Kemp eliminase enzyme KABLE1 with a catalytic efficiency of 7,000 M-1s-1. This activity of KABLE1 was enhanced by about 300-fold over the starting design in only two rounds of directed evolution guided by NMR strategy.1 NMR-guided evolution approach was previously used to evolve a non-enzymatic, heme-binding protein myoglobin to a very highly efficient Kemp eliminase FerrElCat (Ferrous Kemp Elimination Catalyst) with an unprecedented 62,000-fold improvement in catalytic efficiency.1 The enzymatic efficiency of the most evolved KABLE variant, KABLE2.6 is about an order of magnitude higher than the best base-catalyzed Kemp eliminase reported to date, GNCA4 V4-4, a computationally optimized Precambrian beta-lactamase for Kemp elimination.2,3 From the practical point of view, our results drastically expand the possibilities for evolving enzymes to promote novel chemical transformations to design sustainable biocatalysts. From the fundamental standpoint, our work illuminates how the evolution of novel protein functions can emerge from existing proteins. Additionally, our results not only point to the importance of exploring chemical and sequence space simultaneously for rapid evolution for a specific function but also highlight the role of protein dynamics in enzyme evolution.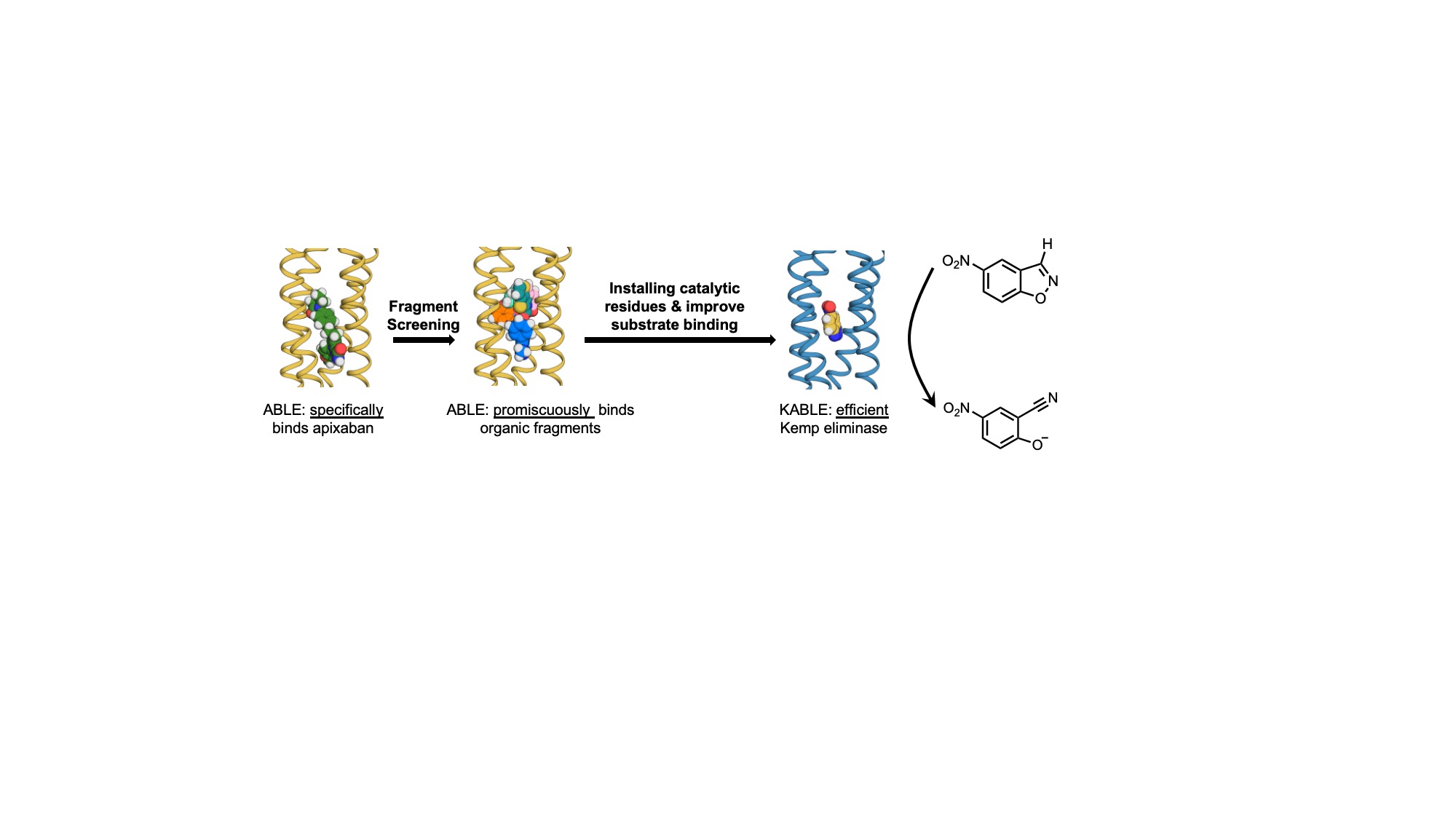
- (1) Bhattacharya, S.; Margheritis, E. G.; Takahashi, K.; Kulesha, A.; D'Souza, A.; Kim, I.; Yoon, J. H.; Tame, J. R. H.; Volkov, A. N.; Makhlynets, O. V.; Korendovych, I. V. NMR-guided directed evolution. Nature 2022, 610, 389-393.
- (2) Risso, V. A.; Martinez-Rodriguez, S.; Candel, A. M.; Kruger, D. M.; Pantoja-Uceda, D.; Ortega-Munoz, M.; Santoyo-Gonzalez, F.; Gaucher, E. A.; Kamerlin, S. C. L.; Bruix, M.; et al. De novo active sites for resurrected Precambrian enzymes. Nat. Commun. 2017, 8, 16113.
- (3) Gutierrez-Rus, L. I.; Vos, E.; Pantoja-Uceda, D.; Hoffka, G.; Gutierrez-Cardenas, J.; Ortega-Muñoz, M.; Risso, V. A.; Jimenez, M. A.; Kamerlin, S. C. L.; Sanchez-Ruiz, J. M. Enzyme Enhancement Through Computational Stability Design Targeting NMR-Determined Catalytic Hotspots. J. Am. Chem. Soc. 2025.